The PSCBS package implements the parent-specific copy-number segmentation presented in Olshen et al. (2011). Package vignette ‘Parent-specific copy-number segmentation using Paired PSCBS’ provides a detailed introduction for running PSCBS segmentation. It’s available as:
Below is an excerpt of the example found in that vignette:
library(PSCBS)
## Get single-chromosome example data
data <- exampleData("paired.chr01")
str(data)
# ’data.frame’: 73346 obs. of 6 variables:
# $ chromosome: int 1 1 1 1 1 1 1 1 1 1 ...
# $ x : int 1145994 2224111 2319424 2543484 2926730 2941694 3084986 3155127..
# $ CT : num 1.625 1.071 1.406 1.18 0.856 ...
# $ betaT : num 0.757 0.771 0.834 0.778 0.229 ...
# $ CN : num 2.36 2.13 2.59 1.93 1.71 ...
# $ betaN : num 0.827 0.875 0.887 0.884 0.103 ...
## Drop total copy-number outliers
data <- dropSegmentationOutliers(data)
## Identify chromosome arms from data
gaps <- findLargeGaps(data, minLength = 1e+06)
knownSegments <- gapsToSegments(gaps)
## Parent-specific copy-number segmentation
fit <- segmentByPairedPSCBS(data, knownSegments = knownSegments)
## Get segments as a data.frame
segments <- getSegments(fit, simplify = TRUE)
segments
# chromosome tcnId dhId start end tcnNbrOfLoci tcnMean
# 1 1 1 1 554484 33414619 9413 1.381375
# 2 1 1 2 33414619 86993745 17433 1.378570
# 3 1 2 1 86993745 87005243 2 3.185100
# 4 1 3 1 87005243 119796080 10404 1.389763
# 5 1 3 2 119796080 119932126 72 1.470789
# 6 1 3 3 119932126 120992603 171 1.439620
# 7 1 4 1 120992604 141510002 0 NA
# 8 1 5 1 141510003 185527989 13434 2.065400
# 9 1 6 1 185527989 199122066 4018 2.707400
# 10 1 7 1 199122066 206512702 2755 2.586100
# 11 1 8 1 206512702 206521352 14 3.871900
# 12 1 9 1 206521352 247165315 15581 2.637500
# tcnNbrOfSNPs tcnNbrOfHets dhNbrOfLoci dhMean c1Mean c2Mean
# 1 2765 2765 2765 0.4868 0.3544608 1.0269140
# 2 4544 4544 4544 0.5185 0.3318907 1.0466792
# 3 0 0 0 NA NA NA
# 4 2777 2777 2777 0.5203 0.3333347 1.0564285
# 5 8 8 8 0.0767 0.6789900 0.7917995
# 6 52 52 52 0.5123 0.3510514 1.0885688
# 7 0 0 NA NA NA NA
# 8 3770 3770 3770 0.0943 0.9353164 1.1300836
# 9 1271 1271 1271 0.2563 1.0067467 1.7006533
# 10 784 784 784 0.2197 1.0089669 1.5771331
# 11 9 9 9 0.2769 1.3998854 2.4720146
# 12 4492 4492 4492 0.2290 1.0167563 1.6207438
## Plot copy-number tracks
plotTracks(fit)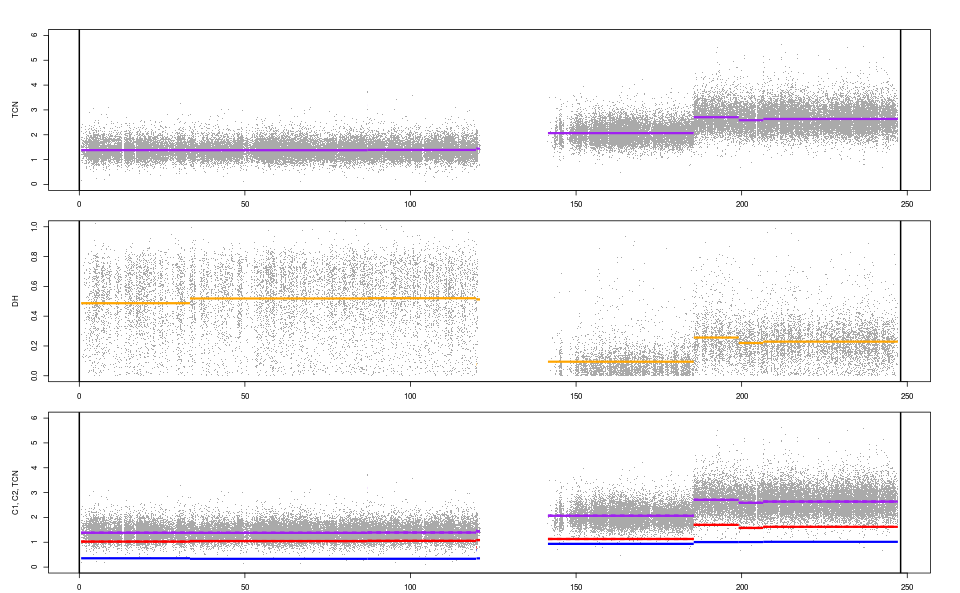
The package supports segmentation of the chromosomes in parallel (asynchronously) via futures by adding the following
to the beginning of the PSCBS script. Everything else will work the same. To reset to non-parallel processing, use future::plan("sequential").
To configure this automatically whenever the package is loaded, see future vignette ‘A Future for R: Controlling Default Future Strategy’.
Bengtsson H, Neuvial P, Speed TP. TumorBoost: Normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays, BMC Bioinformatics, 2010. DOI: 10.1186/1471-2105-11-245. PMID: 20462408, PMCID: PMC2894037
Olshen AB, Bengtsson H, Neuvial P, Spellman PT, Olshen RA, Seshan VA. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation, Bioinformatics, 2011. DOI: 10.1093/bioinformatics/btr329. PMID: 21666266. PMCID: PMC3137217
R package PSCBS is available on CRAN and can be installed in R as:
# install.packages("BiocManager")
BiocManager::install(c("aroma.light", "DNAcopy"))
install.packages("PSCBS")To install the pre-release version that is available in Git branch develop on GitHub, use:
This will install the package from source.