The goal of did2s is to estimate TWFE models without running into the problem of staggered treatment adoption.
For common issues, see this issue: https://github.com/kylebutts/did2s/issues/12
You can install did2s from CRAN with:
install.packages("did2s")To install the development version, run the following:
devtools::install_github("kylebutts/did2s")For details on the methodology, view this vignette
To view the documentation, type ?did2s into the
console.
The main function is did2s which estimates the two-stage
did procedure. This function requires the following options:
yname: the outcome variablefirst_stage: formula for first stage, can include fixed
effects and covariates, but do not include treatment variable(s)!second_stage: This should be the treatment variable or
in the case of event studies, treatment variables.treatment: This has to be the 0/1 treatment variable
that marks when treatment turns on for a unit. If you suspect
anticipation, see note above for accounting for this.cluster_var: Which variables to cluster onOptional options:
weights: Optional variable to run a weighted first- and
second-stage regressionsbootstrap: Should standard errors be bootstrapped
instead? Default is False.n_bootstraps: How many clustered bootstraps to perform
for standard errors. Default is 250.did2s returns a list with two objects:
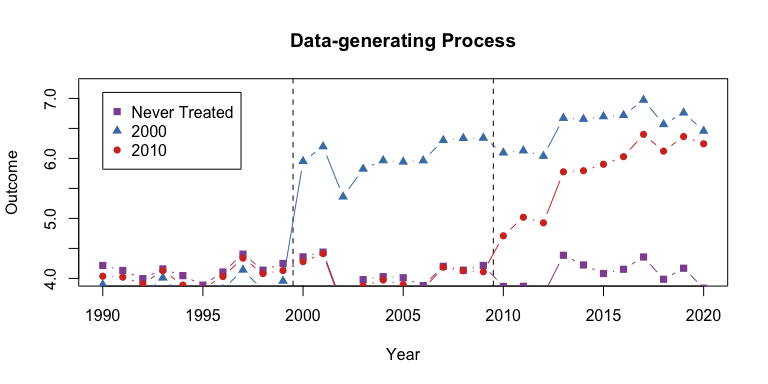
I will load example data from the package and plot the average outcome among the groups.
# Automatically loads fixest
library(did2s)
#> Loading required package: fixest
#> ℹ did2s (v0.7.0). For more information on the methodology, visit <https://www.kylebutts.com/did2s>
#> To cite did2s in publications use:
#>
#> Butts, Kyle (2021). did2s: Two-Stage Difference-in-Differences
#> Following Gardner (2021). R package version 0.7.0.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {did2s: Two-Stage Difference-in-Differences Following Gardner (2021)},
#> author = {Kyle Butts},
#> year = {2021},
#> url = {https://github.com/kylebutts/did2s/},
#> }
# Load Data from R package
data("df_het", package = "did2s")Here is a plot of the average outcome variable for each of the groups:
# Mean for treatment group-year
agg <- aggregate(df_het$dep_var, by=list(g = df_het$g, year = df_het$year), FUN = mean)
agg$g <- as.character(agg$g)
agg$g <- ifelse(agg$g == "0", "Never Treated", agg$g)
never <- agg[agg$g == "Never Treated", ]
g1 <- agg[agg$g == "2000", ]
g2 <- agg[agg$g == "2010", ]
plot(0, 0, xlim = c(1990,2020), ylim = c(4,7.2), type = "n",
main = "Data-generating Process", ylab = "Outcome", xlab = "Year")
abline(v = c(1999.5, 2009.5), lty = 2)
lines(never$year, never$x, col = "#8e549f", type = "b", pch = 15)
lines(g1$year, g1$x, col = "#497eb3", type = "b", pch = 17)
lines(g2$year, g2$x, col = "#d2382c", type = "b", pch = 16)
legend(x=1990, y=7.1, col = c("#8e549f", "#497eb3", "#d2382c"),
pch = c(15, 17, 16),
legend = c("Never Treated", "2000", "2010"))
First, lets estimate a static did. There are two things to note here.
First, note that I can use fixest::feols formula including
the | for specifying fixed effects and
fixest::i for improved factor variable support. Second,
note that did2s returns a fixest estimate
object, so fixest::etable, fixest::coefplot,
and fixest::iplot all work as expected.
# Static
static <- did2s(df_het,
yname = "dep_var", first_stage = ~ 0 | state + year,
second_stage = ~i(treat, ref=FALSE), treatment = "treat",
cluster_var = "state")
#> Running Two-stage Difference-in-Differences
#> • first stage formula `~ 0 | state + year`
#> • second stage formula `~ i(treat, ref = FALSE)`
#> • The indicator variable that denotes when treatment is on is `treat`
#> • Standard errors will be clustered by `state`
fixest::etable(static)
#> static
#> Dependent Var.: dep_var
#>
#> treat = TRUE 2.152*** (0.0476)
#> _______________ _________________
#> S.E. type Custom
#> Observations 46,500
#> R2 0.33790
#> Adj. R2 0.33790
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1This is very close to the true treatment effect of ~2.23.
Then, let’s estimate an event study did. Note that relative year has
a value of Inf for never treated, so I put this as a
reference in the second stage formula.
# Event Study
es <- did2s(df_het,
yname = "dep_var", first_stage = ~ 0 | state + year,
second_stage = ~i(rel_year, ref=c(-1, Inf)), treatment = "treat",
cluster_var = "state")
#> Running Two-stage Difference-in-Differences
#> • first stage formula `~ 0 | state + year`
#> • second stage formula `~ i(rel_year, ref = c(-1, Inf))`
#> • The indicator variable that denotes when treatment is on is `treat`
#> • Standard errors will be clustered by `state`And plot the results:
fixest::iplot(es, main = "Event study: Staggered treatment", xlab = "Relative time to treatment", col = "steelblue", ref.line = -0.5)
# Add the (mean) true effects
true_effects = head(tapply((df_het$te + df_het$te_dynamic), df_het$rel_year, mean), -1)
points(-20:20, true_effects, pch = 20, col = "black")
# Legend
legend(x=-20, y=3, col = c("steelblue", "black"), pch = c(20, 20),
legend = c("Two-stage estimate", "True effect"))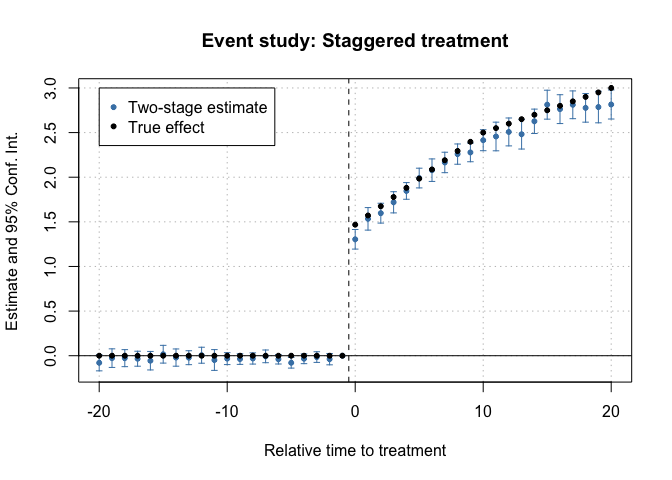
twfe = feols(dep_var ~ i(rel_year, ref=c(-1, Inf)) | unit + year, data = df_het)
fixest::iplot(list(es, twfe), sep = 0.2, ref.line = -0.5,
col = c("steelblue", "#82b446"), pt.pch = c(20, 18),
xlab = "Relative time to treatment",
main = "Event study: Staggered treatment (comparison)")
# Legend
legend(x=-20, y=3, col = c("steelblue", "#82b446"), pch = c(20, 18),
legend = c("Two-stage estimate", "TWFE"))
If you use this package to produce scientific or commercial publications, please cite according to:
citation(package = "did2s")
#>
#> To cite did2s in publications use:
#>
#> Butts, Kyle (2021). did2s: Two-Stage Difference-in-Differences
#> Following Gardner (2021). R package version 0.7.0.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {did2s: Two-Stage Difference-in-Differences Following Gardner (2021)},
#> author = {Kyle Butts},
#> year = {2021},
#> url = {https://github.com/kylebutts/did2s/},
#> }