The purpose of dvir is to implement state-of-the-art algorithms for DNA-based disaster victim identification (DVI). In particular, dvir performs joint identification of multiple victims.
The methodology and algorithms of dvir are described in Vigeland & Egeland (2021): DNA-based Disaster Victim Identification.
The dvir package is part of the ped suite, a collection of R packages for relatedness pedigree analysis. Much of the machinery behind dvir is imported from other ped suite packages, especially pedtools for handling pedigrees and marker data, and forrel for the calculation of likelihood ratios. A comprehensive presentation of these packages, and much more, can be found in the recently published book Pedigree Analysis in R.
To get the current official version of dvir, install from CRAN as follows:
Alternatively, the latest development version may be obtained from GitHub:
# First install devtools if needed
if(!require(devtools)) install.packages("devtools")
# Install dvir from GitHub
devtools::install_github("thoree/dvir")In the following we will use a toy DVI example from the paper (see above) to illustrate how to use dvir.
To get started, we load the dvir package.
We consider the DVI problem shown below, in which three victim samples (V1, V2, V3) are to be matched against three missing persons (M1, M2, M3) belonging to two different families.
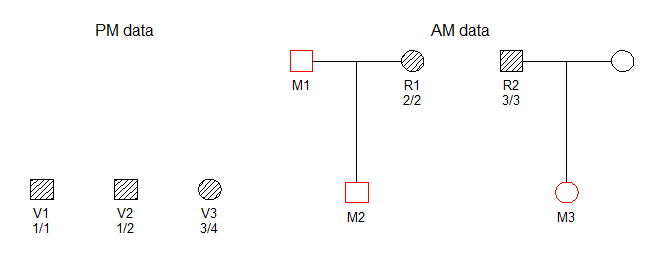
The hatched symbols indicate genotyped individuals. In this simple example we consider only a single marker, with 10 equifrequent alleles denoted 1, 2,…, 10. The available genotypes are shown in the figure.
DNA profiles from victims are generally referred to as post mortem (PM) data, while the ante mortem (AM) data contains profiles from the reference individuals R1 and R2.
A possible solution to the DVI problem is called an assignment. In our toy example, there are a priori 14 possible assignments, which can be listed as follows:
#> V1 V2 V3
#> 1 M1 M2 M3
#> 2 M1 M2 *
#> 3 * M2 M3
#> 4 M1 * M3
#> 5 * M1 M3
#> 6 * M2 *
#> 7 * * M3
#> 8 M1 * *
#> 9 * M1 *
#> 10 * * *
#> 11 M2 M1 M3
#> 12 M2 M1 *
#> 13 M2 * M3
#> 14 M2 * *Each row indicates the missing persons corresponding to V1, V2 and V3 (in that order) with * meaning not identified. For example, the first line gives the assignment where (V1, V2, V3) = (M1, M2, M3), while line 10 shows the null model corresponding to none of the victims identified. For each assignment a we can calculate the likelihood, denoted L(a). The null likelihood is denoted L0.
We consider the following to be the two main goals of the DVI analysis:
a to the null model: LR = L(a)/L0.P(Vi = Mj | data) for all combinations of i,j = 1,2,3, and the posterior non-pairing probabilities P(Vi = '*' | data) for all i = 1, 2, 3.The genotypes for this toy example are available within dvir as a built-in dataset, under the name example2. This has the structure of a list, with elements pm (the victim data), am (the reference data) and missing (a vector naming the missing persons). For easy reference we store them in separate variables.
We can inspect the data by printing each object. For instance, am is a list of two pedigrees:
am
#> [[1]]
#> id fid mid sex L1
#> M1 * * 1 -/-
#> R1 * * 2 2/2
#> M2 M1 R1 1 -/-
#>
#> [[2]]
#> id fid mid sex L1
#> R2 * * 1 3/3
#> MO2 * * 2 -/-
#> M3 R2 MO2 2 -/-Note that the two pedigrees are printed in so-called ped format, with columns id (ID label), fid (father), mid (mother), sex (sex coded as 1 = male; 2 = female) and L1 (genotypes at locus L1).
The appendix contains code for generating this dataset from scratch.
The jointDVI() function performs joint identification of all three victims, given the data. It returns a data frame ranking all assignments with nonzero likelihood:
jointRes = jointDVI(pm, am, missing, verbose = FALSE)
# Print the result
jointRes
#> V1 V2 V3 loglik LR posterior
#> 1 M1 M2 M3 -16.11810 250 0.718390805
#> 2 M1 M2 * -17.72753 50 0.143678161
#> 3 * M2 M3 -18.42068 25 0.071839080
#> 4 M1 * M3 -20.03012 5 0.014367816
#> 5 * M1 M3 -20.03012 5 0.014367816
#> 6 * M2 * -20.03012 5 0.014367816
#> 7 * * M3 -20.03012 5 0.014367816
#> 8 M1 * * -21.63956 1 0.002873563
#> 9 * M1 * -21.63956 1 0.002873563
#> 10 * * * -21.63956 1 0.002873563The output shows that the most likely joint solution is (V1, V2, V3) = (M1, M2, M3), with an LR of 250 compared to the null model.
Next, we compute the posterior pairing (and non-pairing) probabilities. This is done by feeding the output from jointDVI() into the function Bmarginal().
Bmarginal(jointRes, missing, prior = NULL)
#> M1 M2 M3 *
#> V1 0.87931034 0.0000000 0.0000000 0.12068966
#> V2 0.01724138 0.9482759 0.0000000 0.03448276
#> V3 0.00000000 0.0000000 0.8333333 0.16666667Here we used a default flat prior for simplicity, assigning equal prior probabilities to all assignments.
we see that the posterior pairing probabilities for the most likely solution are
For completeness, here is one way of generating the example2 dataset from scratch within R.
# Load pedtools for pedigree manipulation
library(pedtools)
# Attributes of the marker locus
loc = list(name = "L1",
alleles = 1:10,
afreq = rep(1/10, 10))
### 1. PM data
# PM as data frame
pm.df = data.frame(famid = c("V1", "V2", "V3"),
id = c("V1", "V2", "V3"),
fid = c(0, 0, 0),
mid = c(0, 0, 0),
sex = c(1, 1, 2),
L1 = c("1/1", "1/2", "3/4"))
# Convert to list of singletons
pm = as.ped(pm.df, locusAttributes = loc)
### 2. AM data
# List of two pedigrees
am = list(nuclearPed(father = "M1", mother = "R1", child = "M2"),
nuclearPed(father = "R2", mother = "MO2", child = "M3", sex = 2))
# Attach marker and set genotypes
am = setMarkers(am, locusAttributes = loc)
genotype(am[[1]], marker = "L1", id = "R1") = "2/2"
genotype(am[[2]], marker = "L1", id = "R2") = "3/3"
### 3. Missing persons
missing = c("M1", "M2", "M3")The complete dataset now looks as follows.
list(pm = pm, am = am, missing = missing)
#> $pm
#> $pm$V1
#> id fid mid sex L1
#> V1 * * 1 1/1
#>
#> $pm$V2
#> id fid mid sex L1
#> V2 * * 1 1/2
#>
#> $pm$V3
#> id fid mid sex L1
#> V3 * * 2 3/4
#>
#>
#> $am
#> $am[[1]]
#> id fid mid sex L1
#> M1 * * 1 -/-
#> R1 * * 2 2/2
#> M2 M1 R1 1 -/-
#>
#> $am[[2]]
#> id fid mid sex L1
#> R2 * * 1 3/3
#> MO2 * * 2 -/-
#> M3 R2 MO2 2 -/-
#>
#>
#> $missing
#> [1] "M1" "M2" "M3"