The goal of ensModelVis is to display model fits for multiple models and their ensembles.
You can install the development version of ensModelVis from GitHub with:
# install.packages("devtools")
devtools::install_github("domijan/ensModelVis")This is a basic example:
library(ensModelVis)
data(iris)
if (require("MASS")) {
lda.model <- lda(Species ~ ., data = iris)
lda.pred <- predict(lda.model)
}
#> Loading required package: MASS
if (require("ranger")) {
ranger.model <- ranger(Species ~ ., data = iris, mtry = 1)
ranger.pred <- predict(ranger.model, iris)
ranger.model2 <-
ranger(Species ~ .,
data = iris,
mtry = 4,
num.trees = 10)
ranger.pred2 <- predict(ranger.model2, iris)
}
#> Loading required package: ranger
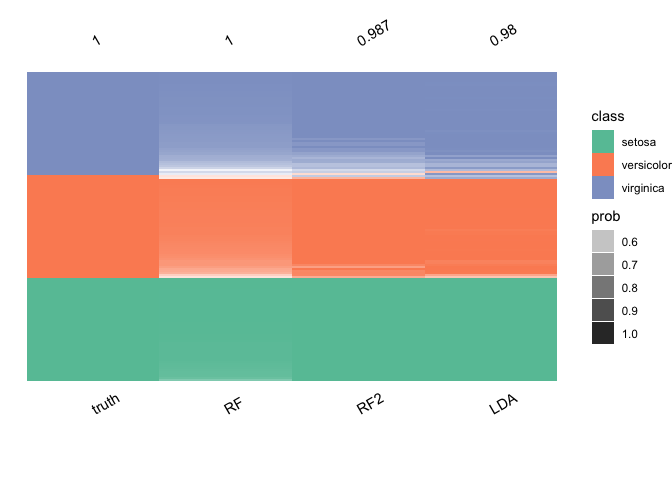
plot_ensemble(
iris$Species,
data.frame(
LDA = lda.pred$class,
RF = ranger.pred$predictions,
RF2 = ranger.pred2$predictions
)
)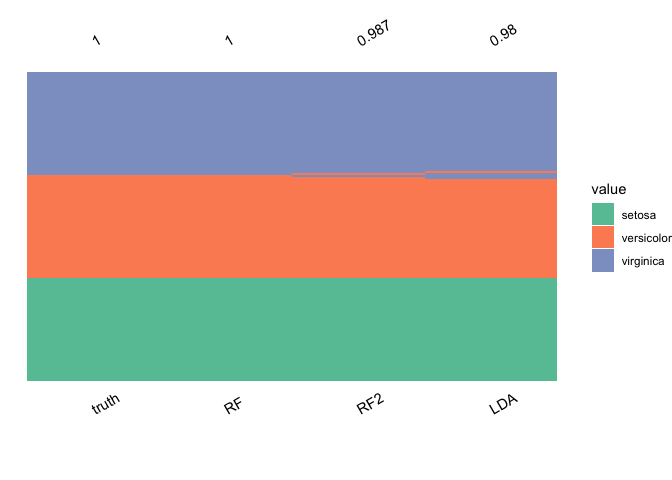
plot_ensemble(
iris$Species,
data.frame(
LDA = lda.pred$class,
RF = ranger.pred$predictions,
RF2 = ranger.pred2$predictions
),
incorrect = TRUE
)
if (require("ranger")) {
ranger.model <- ranger(Species ~ ., data = iris, mtry = 1, probability = TRUE)
ranger.prob <- predict(ranger.model, iris)
ranger.model2 <-
ranger(Species ~ .,
data = iris,
mtry = 4,
num.trees = 10,
probability = TRUE)
ranger.prob2 <- predict(ranger.model2, iris)
}
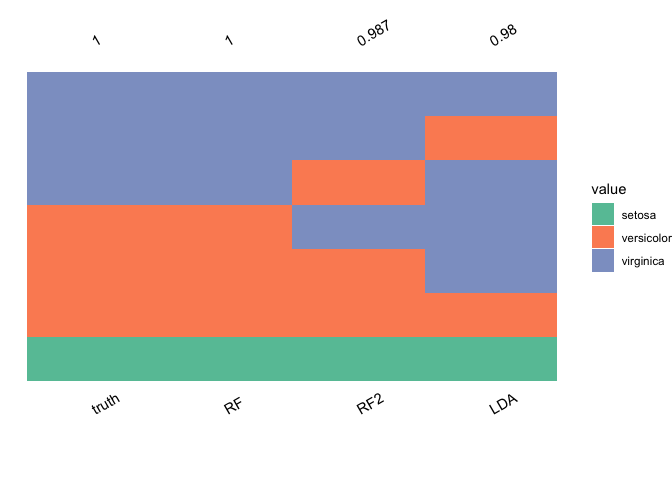
plot_ensemble(
iris$Species,
data.frame(LDA = lda.pred$class,
RF = ranger.pred$predictions,
RF2 = ranger.pred2$predictions),
tibble_prob = data.frame(
LDA = apply(lda.pred$posterior, 1, max),
RF = apply(ranger.prob$predictions, 1, max),
RF2 = apply(ranger.prob2$predictions, 1, max)
)
)