The package supports shelf life estimation for chemically derived medicines, either following the standard method proposed by the International Council for Harmonisation (ICH), in quality guideline Q1E Evaluation of Stability Data) or following the worst-case scenario consideration (what-if analysis) described in the Australian Regulatory Guidelines for Prescription Medicines (ARGPM), guidance on (Stability testing for prescription medicines).
A stable version of expirest can be installed from
CRAN:
# install.packages("expirest")The development version is available from GitHub by:
# install.packages("devtools")
devtools::install_github("piusdahinden/expirest")This is a basic example which shows you how to solve a common problem using a data set containing the moisture stability data (% (w/w)) of three batches obtained over a 24 months period of a drug product. A total of n = 33 independent measurements are available (corresponding to data shown in Table XIII in LeBlond et al. (2011)).
library(expirest)
# Data frame
str(exp3)
#> 'data.frame': 33 obs. of 3 variables:
#> $ Batch : Factor w/ 3 levels "b1","b2","b3": 1 1 1 1 1 1 1 1 1 1 ...
#> $ Month : num 0 1 2 3 3 6 6 12 12 24 ...
#> $ Moisture: num 2.2 1.7 3.32 2.76 2.43 ...
# Perform what-if shelf life estimation (wisle) and print a summary
res1 <- expirest_wisle(
data = exp3, response_vbl = "Moisture", time_vbl = "Month",
batch_vbl = "Batch", rl = 3.00, rl_sf = 3, sl = c(0.5, 4.5),
sl_sf = c(1, 2), srch_range = c(0, 500), alpha = 0.05,
alpha_pool = 0.25, xform = c("no", "no"), shift = c(0, 0),
sf_option = "tight", ivl = "confidence", ivl_type = "one.sided",
ivl_side = "upper")
class(res1)
#> [1] "expirest_wisle"
summary(res1)
#>
#> Summary of shelf life estimation following the ARGPM
#> guidance "Stability testing for prescription medicines"
#>
#> The best model accepted at a significance level of 0.25 has
#> Common intercepts and Common slopes (acronym: cics).
#>
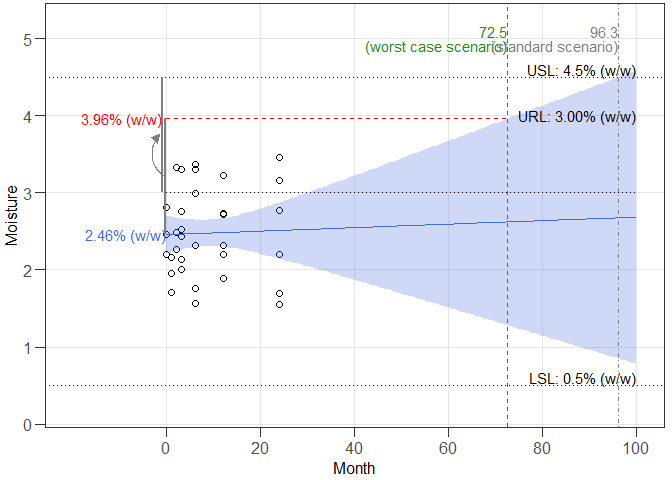
#> Worst case intercept(s): 2.456782
#> Worst case batch(es): NA
#>
#> Estimated shelf life (lives) for cics model:
#> SL RL wisle osle
#> cics 4.5 3 72.50545 96.30552
#>
#> Abbreviations:
#> ARGPM: Australian Regulatory Guidelines for Prescription Medicines; ICH: International Council for Harmonisation; osle: Ordinary shelf life estimation (i.e. following the ICH guidance); RL: Release Limit; SL: Specification Limit; wisle: What-if (approach for) shelf life estimation (i.e. following ARGPM guidance).
# Prepare graphical representation
ggres1 <- plot_expirest_wisle(
model = res1, rl_index = 1, show_grouping = "no",
response_vbl_unit = "% (w/w)", y_range = c(0.2, 5.2),
x_range = NULL, scenario = "standard", plot_option = "full",
ci_app = "ribbon")
class(ggres1)
#> [1] "plot_expirest_wisle"
plot(ggres1)
The model type in Example 1 was common intercept / common slope (cics). The model type in this example is different intercept / different slope (dids). A data set containing the potency stability data (in % of label claim (LC)) of five batches of a drug product obtained over a 24 months period is used. A total of n = 53 independent measurements are available (corresponding to data shown in Tables IV, VI and VIII in LeBlond et al. (2011)).
library(expirest)
# Data frame
str(exp1)
#> 'data.frame': 53 obs. of 3 variables:
#> $ Batch : Factor w/ 6 levels "b2","b3","b4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ Month : num 0 1 3 3 6 6 12 12 24 24 ...
#> $ Potency: num 101 101.3 99.8 99.2 99.5 ...
# Perform what-if shelf life estimation (wisle) and print a summary
res1 <- expirest_wisle(
data = exp1[exp1$Batch %in% c("b4", "b5", "b8"), ],
response_vbl = "Potency", time_vbl = "Month", batch_vbl = "Batch",
rl = c(98.0, 98.5, 99.0), rl_sf = rep(2, 3), sl = 95, sl_sf = 2,
srch_range = c(0, 500), alpha = 0.05, alpha_pool = 0.25,
xform = c("no", "no"), shift = c(0, 0), sf_option = "tight",
ivl = "confidence", ivl_type = "one.sided", ivl_side = "lower")
summary(res1)
#>
#> Summary of shelf life estimation following the ARGPM
#> guidance "Stability testing for prescription medicines"
#>
#> The best model accepted at a significance level of 0.25 has
#> Different intercepts and Different slopes (acronym: dids).
#>
#> Worst case intercept(s): 101.2594 101.2594 101.2594
#> Worst case batch(es): b8 b8 b8
#>
#> Estimated shelf life (lives) for dids model:
#> SL RL wisle osle
#> 1 95 98.0 7.619661 15.84487
#> 2 95 98.5 8.997036 15.84487
#> 3 95 99.0 10.303030 15.84487
#>
#> Abbreviations:
#> ARGPM: Australian Regulatory Guidelines for Prescription Medicines; ICH: International Council for Harmonisation; osle: Ordinary shelf life estimation (i.e. following the ICH guidance); RL: Release Limit; SL: Specification Limit; wisle: What-if (approach for) shelf life estimation (i.e. following ARGPM guidance).
# Prepare graphical representation
ggres1 <- plot_expirest_wisle(
model = res1, rl_index = 2, show_grouping = "yes",
response_vbl_unit = "%", y_range = c(93, 107),
x_range = NULL, scenario = "standard", plot_option = "full",
ci_app = "ribbon")
class(ggres1)
#> [1] "plot_expirest_wisle"
plot(ggres1)
#> Warning: Removed 4 rows containing missing values (geom_point).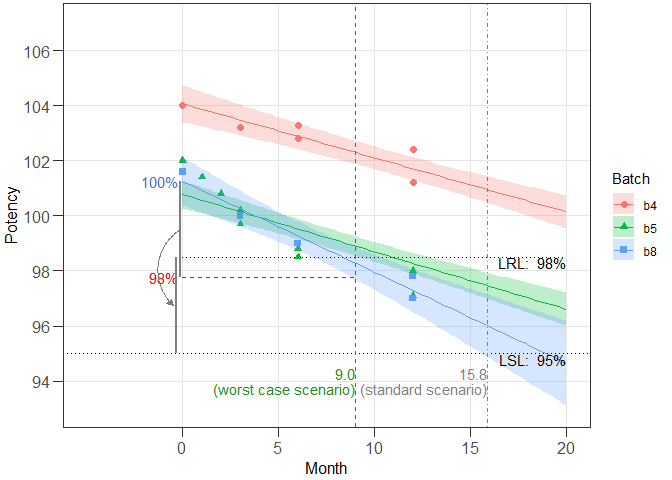
Pius Dahinden, Tillotts Pharma AG