

The fwildclusterboot package provides a native R
implementation of the fast wild cluster bootstrap algorithm developed in
Roodman
et al (2019) for regression objects in R.
It also ports functionality of WildBootTests.jl to R via the JuliaConnectoR.
The package’s central function is boottest(). It allows
to test univariate hypotheses using a wild cluster bootstrap at extreme
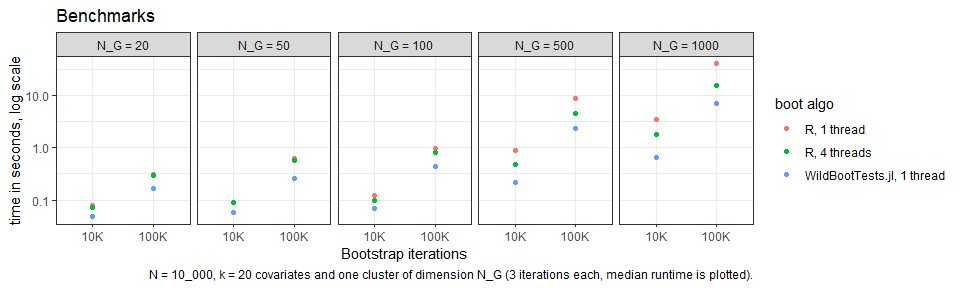
speed: via the ‘fast’ algorithm, it is possible to run a wild cluster
bootstrap with iterations in less than a
second!
fwildclusterboot supports the following features:
Additional features are provided through
WildBootTests.jl:
fwildclusterboot supports the following models:
lm (from stats), fixest (from
fixest), felm from (lfe)ivreg (from ivreg).boottest() functionFor a longer introduction to fwildclusterboot, take a
look at the vignette.
library(fwildclusterboot)
# set seed via dqset.seed for boot_algo = "R" & Rademacher, Webb & Normal weights
dqrng::dqset.seed(2352342)
# set 'familiar' seed for all other algorithms and weight types
set.seed(23325)
data(voters)
# fit the model via fixest::feols(), lfe::felm() or stats::lm()
lm_fit <- lm(proposition_vote ~ treatment + log_income + as.factor(Q1_immigration) + as.factor(Q2_defense), data = voters)
# bootstrap inference via boottest()
lm_boot <- boottest(lm_fit, clustid = c("group_id1"), B = 9999, param = "treatment", seed = 1)
summary(lm_boot)
#> boottest.lm(object = lm_fit, param = "treatment", B = 9999, clustid = c("group_id1"),
#> seed = 1)
#>
#> Hypothesis: 1*treatment = 0
#> Observations: 300
#> Bootstr. Type: rademacher
#> Clustering: 1-way
#> Confidence Sets: 95%
#> Number of Clusters: 40
#>
#> term estimate statistic p.value conf.low conf.high
#> 1 1*treatment = 0 0.079 3.983 0 0.04 0.118You can install compiled versions offwildclusterboot
from CRAN and the development version from R-universe (compiled) or
github by following one of the steps below:
# from CRAN
install.packages("fwildclusterboot")
# from r-universe (windows & mac, compiled R > 4.0 required)
install.packages('fwildclusterboot', repos ='https://s3alfisc.r-universe.dev')
# dev version from github
# note: installation requires Rtools
library(devtools)
install_github("s3alfisc/fwildclusterboot")To run WildBootTests.jl through
fwildclusterboot, Julia and
WildBootTests.jl need to be installed.
You can install Julia by following the steps described on the
official Julia
homepage. WildBootTests.jl can then be installed via
Julia’s package management system.
To help you connect Julia and R via the JuliaConnectoR
and to install WildBootTests.jl from within R, you can
alternatively use the JuliaConnectoR.utils package.
devtools::install_github("s3alfisc/JuliaConnectoR.utils")
library(JuliaConnectoR.utils)
connect_julia_r() # instructions to connect Julia and R
install_julia_packages("WildBootTests.jl") # install WildBootTests.jl
set_julia_nthreads() # instructions to set nthreads for Julia