{ggblanket} is a package of {ggplot2} wrapper functions to simplify visualisation.
To do this, the {ggblanket} package:
gg_* functions that wrap a single
ggplot2::geom_* functioncol
argumentpal and alpha
arguments consistentlyfacet argument to facet by a single
variablefacet2 argument to facet in a
gridtheme argument for customisation.gg_theme function to create a quick
theme.snakecase::to_sentencegg_blank function for extra flexibilityplotly::ggplotly tooltipsgeom_* arguments via
...If you would like to show your appreciation for {ggblanket}, you can give this repository a star, or even buy me a coffee.
Click here for the {ggblanket} website.
Install either from CRAN with:
install.packages("ggblanket")Or install the development version with:
# install.packages("devtools")
devtools::install_github("davidhodge931/ggblanket")library(dplyr)
library(ggplot2)
library(ggblanket)
library(palmerpenguins)gg_* functions that wrap a single
ggplot2::geom_* function.iris %>%
mutate(Species = stringr::str_to_sentence(Species)) %>%
gg_point(
x = Sepal.Width,
y = Sepal.Length,
col = Species)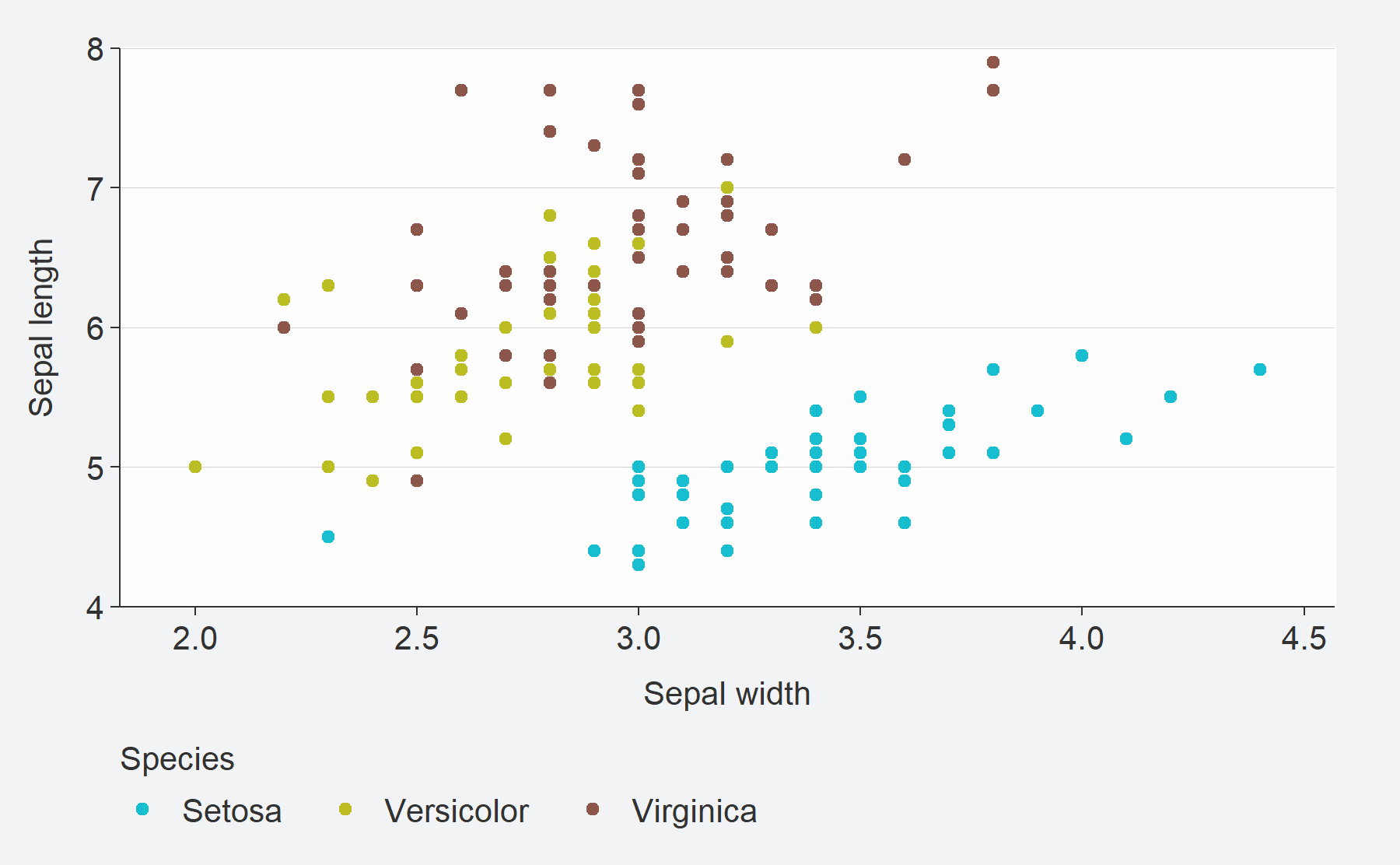
col argument.penguins %>%
gg_histogram(
x = body_mass_g,
col = species) 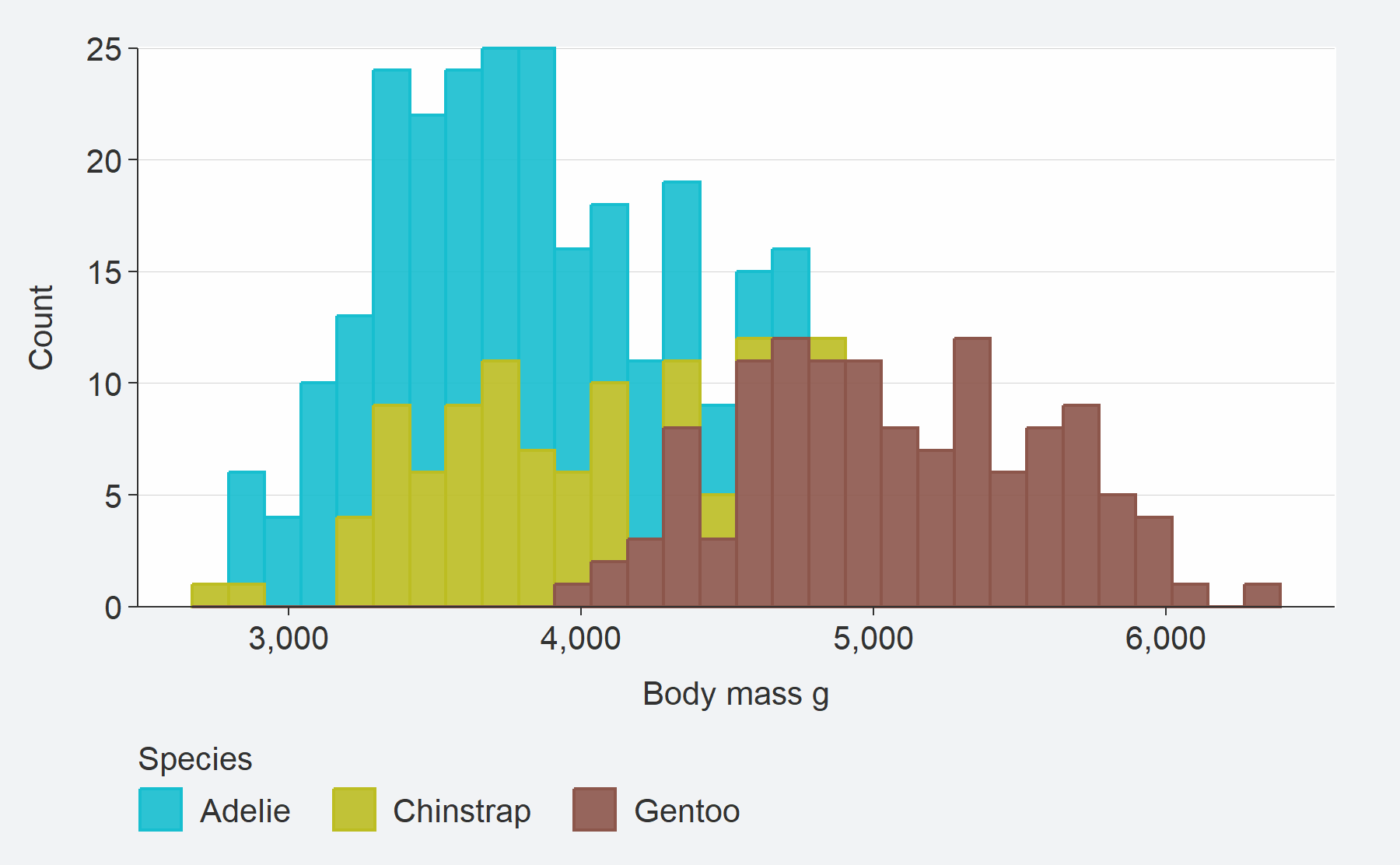
pal and
alpha arguments consistently.These arguments are the same regardless of whether a col
variable is specified. If more colours are provided than needed by the
pal argument, then the excess colours will just be dropped. Note all
colours specified by the pal argument will inherit to any further
ggplot2::geom_* layers added.
penguins %>%
mutate(sex = stringr::str_to_sentence(sex)) %>%
group_by(species, sex) %>%
summarise(body_mass_g = mean(body_mass_g, na.rm = TRUE)) %>%
gg_col(
x = species,
y = body_mass_g,
col = sex,
position = position_dodge2(preserve = "single"),
pal = c("#1B9E77", "#9E361B"))
facet argument to facet by a
single variable.penguins %>%
tidyr::drop_na(sex) %>%
mutate(sex = stringr::str_to_sentence(sex)) %>%
gg_violin(
x = sex,
y = body_mass_g,
facet = species,
y_include = 0,
y_breaks = scales::breaks_width(1000),
pal = "#1B9E77")
facet2 argument to
facet in a grid.penguins %>%
tidyr::drop_na(sex) %>%
mutate(sex = stringr::str_to_sentence(sex)) %>%
gg_point(
x = bill_length_mm,
y = body_mass_g,
col = sex,
facet = species,
facet2 = sex,
y_breaks = scales::breaks_width(1500),
size = 1)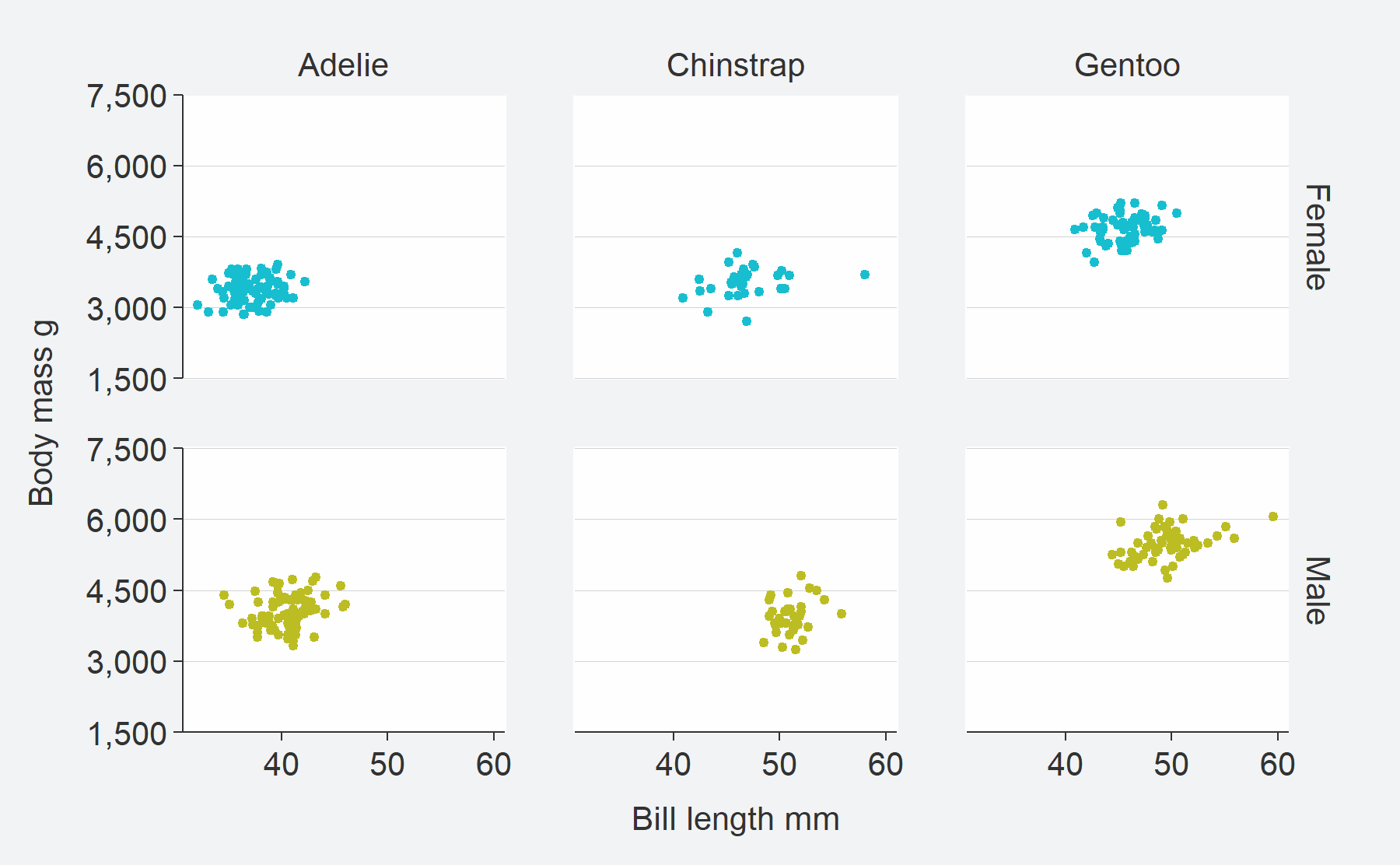
This is designed to work with the Rstudio autocomplete to help you
find the adjustment you need. Press the tab key after typing
x_,y_, col_ or
facet_ to access this. Then use arrow keys, and press tab
again to select.
Available arguments are:
*_breaks: Adjust the breaks of an axis*_expand: Adjust the padding beyond the limits*_include: Include a value within a scale*_labels: Adjust the labels on the breaks*_limits: Adjust the limits*_trans: Transform an axis*_sec_axis: Add a secondary axiscol_intervals: Determine intervals to colour by.penguins %>%
gg_jitter(
x = species,
y = body_mass_g,
col = flipper_length_mm,
position = ggplot2::position_jitter(width = 0.2, height = 0, seed = 123),
col_intervals = ~ santoku::chop_quantiles(.x, probs = seq(0, 1, 0.25)),
col_legend_place = "r",
y_include = 0,
y_breaks = scales::breaks_width(1500),
y_labels = scales::label_number()
)
Where x variable is categorical and y numeric, the numeric y scale defaults to the limits being the min and max of the breaks, with expand of c(0, 0). Equivalent happens for the horizontal vice versa situation.
Where both x and y are numeric/date, the y scale defaults to the
limits being the min and max of the breaks with expand of c(0,
0) - and x scales default to the min and max of the variable
with expand of c(0.025, 0.025).
storms %>%
group_by(year) %>%
filter(between(year, 1980, 2020)) %>%
summarise(wind = mean(wind, na.rm = TRUE)) %>%
gg_line(
x = year,
y = wind,
x_labels = ~.x,
y_include = 0,
title = "Storm wind speed",
subtitle = "USA average storm wind speed, 1980\u20132020",
y_title = "Wind speed (knots)",
caption = "Source: NOAA"
) +
geom_point()
theme argument for
customisation.This allows you to utilise the simplicity of {ggblanket}, while making content that has your required look and feel.
Your theme will control all theme aspects, other than the legend
position and direction. You must instead control these within the
gg_* function with the col_legend_place
argument (e.g. `col_legend_place = "r").
penguins %>%
mutate(sex = stringr::str_to_sentence(sex)) %>%
gg_point(x = bill_depth_mm,
y = bill_length_mm,
col = sex,
facet = species,
pal = c("#1B9E77", "#9E361B"),
theme = theme_grey())
gg_theme function to create a
quick theme.The gg_theme function allows you to create a theme that
looks similar to the {ggblanket} look and feel.
This includes the following arguments for adjusting gridlines, background colours, text and axis lines and ticks.
storms %>%
group_by(year) %>%
filter(between(year, 1980, 2020)) %>%
summarise(wind = mean(wind, na.rm = TRUE)) %>%
gg_col(
x = year,
y = wind,
x_labels = ~.x,
x_expand = c(0, 0),
theme = gg_theme(
bg_plot_pal = "white",
bg_panel_pal = "white",
grid_h = TRUE))
penguins %>%
tidyr::drop_na(sex) %>%
group_by(species, sex, island) %>%
summarise(body_mass_kg = mean(body_mass_g) / 1000) %>%
gg_col(
x = body_mass_kg,
y = species,
col = sex,
facet = island,
col_labels = snakecase::to_sentence_case,
position = "dodge")
penguins %>%
group_by(species, sex) %>%
summarise(across(body_mass_g, ~ round(mean(.x, na.rm = TRUE)), 0)) %>%
gg_tile(
x = sex,
y = species,
col = body_mass_g,
x_labels = snakecase::to_sentence_case,
pal = pals::brewer.blues(9),
width = 0.9,
height = 0.9,
col_legend_place = "r",
title = "Average penguin body mass",
subtitle = "Palmer Archipelago, Antarctica",
theme = gg_theme(grid_h = FALSE,
bg_plot_pal = "white",
axis_pal = "white",
ticks_pal = "white")) +
geom_text(aes(label = body_mass_g), col = "#232323", size = 3.5) 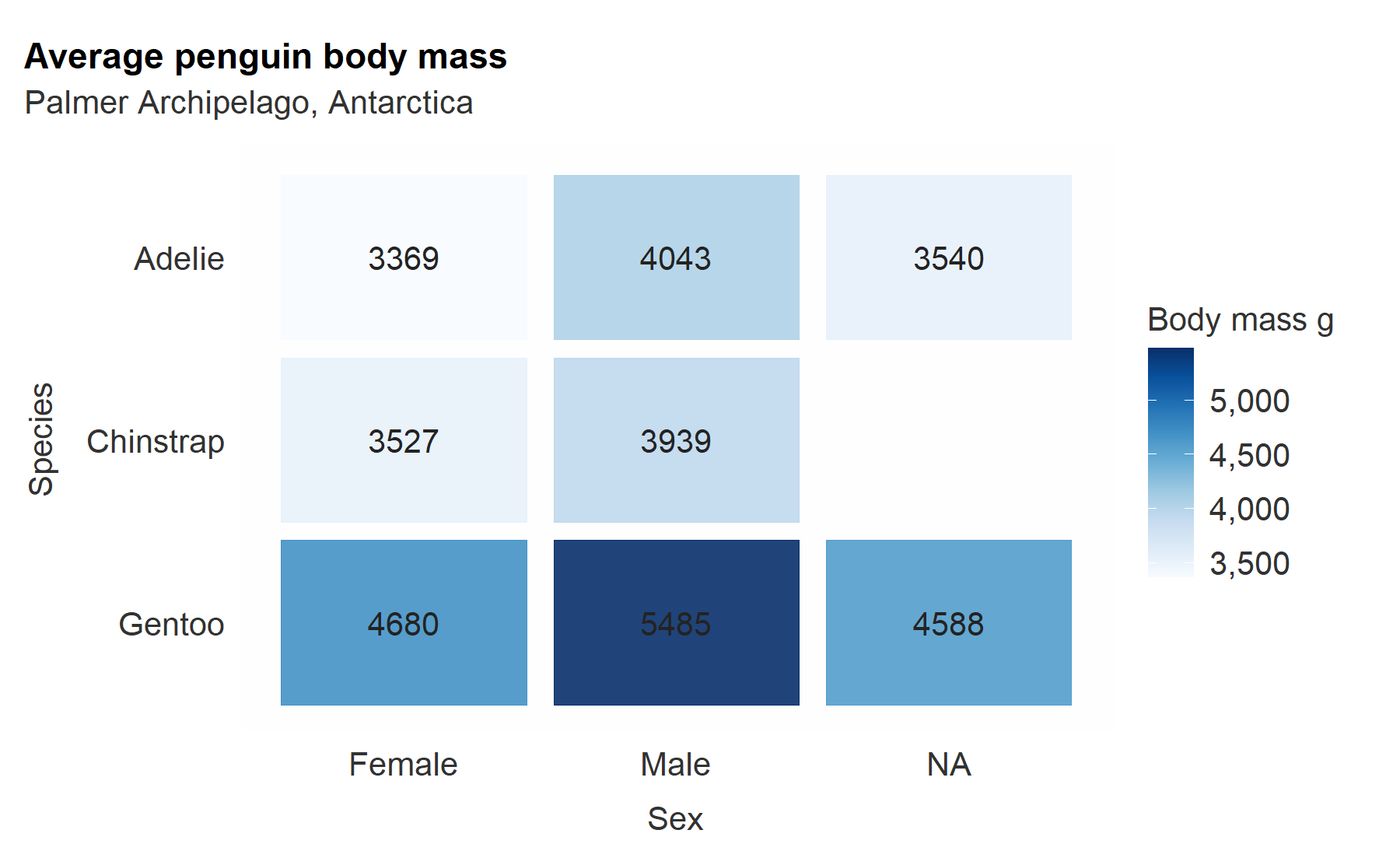
gg_blank function for extra
flexibility.penguins %>%
tidyr::drop_na(sex) %>%
mutate(sex = stringr::str_to_sentence(sex)) %>%
group_by(species, sex) %>%
summarise(
mean = round(mean(bill_length_mm, na.rm = TRUE), 0),
n = n(),
se = mean / sqrt(n),
upper = mean + 1.96 * se,
lower = mean - 1.96 * se
) %>%
gg_blank(
x = sex,
y = mean,
col = sex,
facet = species,
label = mean,
ymin = lower,
ymax = upper,
y_include = 0,
y_title = "Bill length mm"
) +
geom_col(width = 0.75, alpha = 0.9) +
geom_errorbar(width = 0.1, colour = pal_na()) 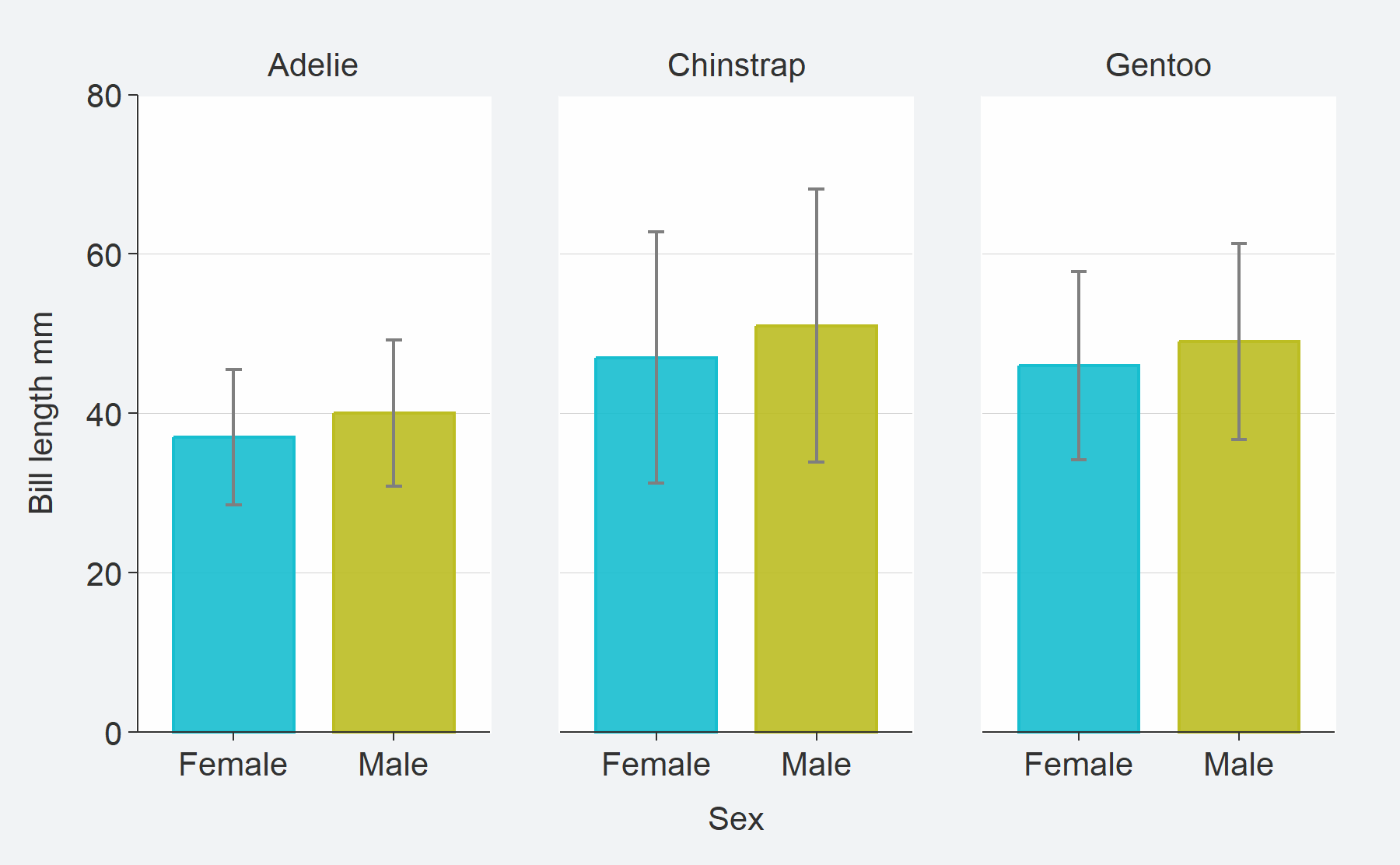
plotly::ggplotly
tooltips.The add_tooltip function allows users to create nice
tooltips in combination with the text argument, and the
tooltip = "text" argument in ggplotly.
theme_custom <- gg_theme(
"helvetica",
bg_plot_pal = "white",
bg_panel_pal = "white",
grid_h = TRUE
)
iris %>%
mutate(Species = stringr::str_to_sentence(Species)) %>%
add_tooltip_text(titles = snakecase::to_sentence_case) %>%
gg_point(
x = Sepal.Width,
y = Sepal.Length,
col = Species,
text = text,
col_legend_place = "r",
theme = theme_custom) %>%
plotly::ggplotly(tooltip = "text")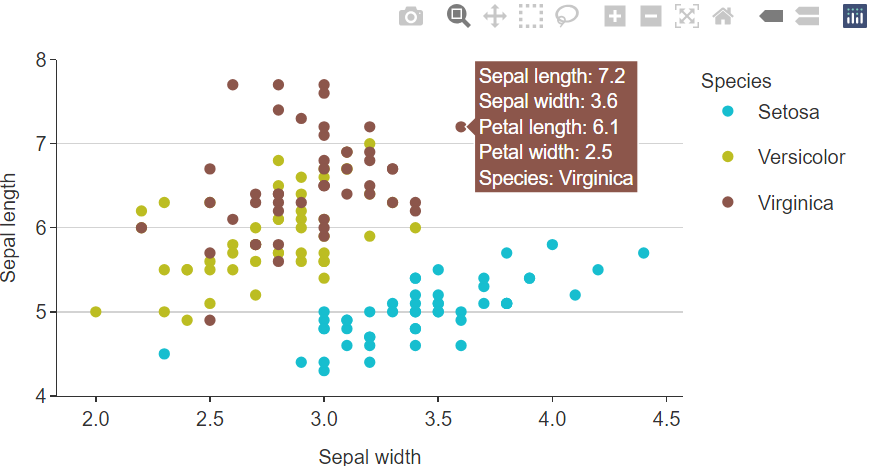
geom_*
arguments via ...penguins %>%
tidyr::drop_na(sex) %>%
gg_smooth(
x = flipper_length_mm,
y = body_mass_g,
col = sex,
level = 0.99, #argument from geom_smooth
col_legend_place = "t",
col_title = "",
col_labels = snakecase::to_sentence_case
) 
This is because the ... argument can allow you to access
all arguments within the {ggblanket} gg_
function.
gg_point_custom <- function(data, x, y, col,
size = 3,
pal = pals::brewer.dark2(9),
col_title = "",
col_legend_place = "t",
...) {
data %>%
gg_point(x = {{ x }}, y = {{ y }}, col = {{col}},
size = size,
pal = pal,
col_title = col_title,
col_legend_place = col_legend_place,
...)
}
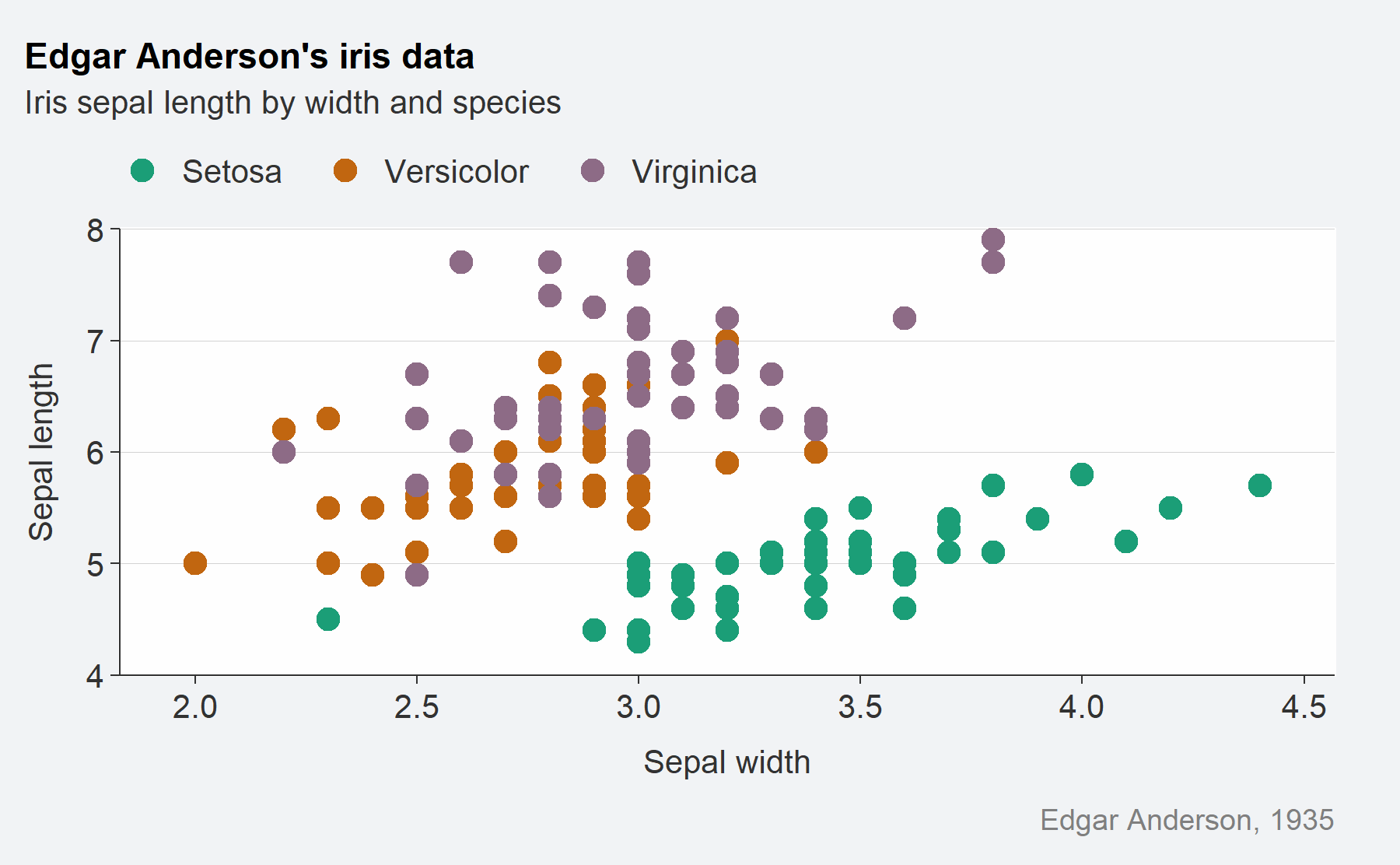
iris %>%
mutate(Species = stringr::str_to_sentence(Species)) %>%
gg_point_custom(
x = Sepal.Width,
y = Sepal.Length,
col = Species,
title = "Edgar Anderson's iris data",
subtitle = "Iris sepal length by width and species",
caption = "Edgar Anderson, 1935"
)