The poems package provides a framework of interoperable R6 (Chang, 2020) classes for building ensembles of viable models via the pattern-oriented modeling (POM) approach (Grimm et al., 2005). The package includes classes for encapsulating and generating model parameters, and managing the POM workflow. The workflow includes:
By default, model validation and selection utilizes an approximate Bayesian computation (ABC) approach (Beaumont et al., 2002) using the abc package (Csillery et al., 2015). However, alternative user-defined functionality could be employed.
The package includes a spatially explicit demographic population model simulation engine, which incorporates default functionality for density dependence, correlated environmental stochasticity, stage-based transitions, and distance-based dispersal. The user may customize the simulator by defining functionality for trans-locations, harvesting, mortality, and other processes, as well as defining the sequence order for the simulator processes. The framework could also be adapted for use with other model simulators by utilizing its extendable (inheritable) base classes.
You can install the released version of poems from CRAN with:
And the development version from GitHub with:
The following simple example demonstrates how to run a single spatially explicit demographic population model using poems:
library(poems)
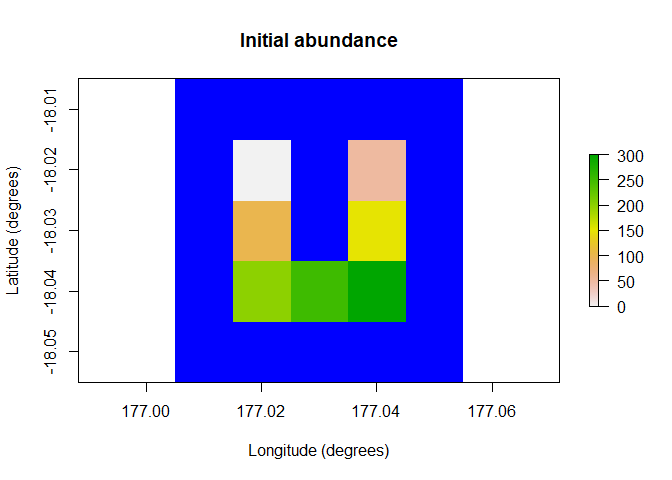
# Demonstration example region (U Island) and initial abundance
coordinates <- data.frame(x = rep(seq(177.01, 177.05, 0.01), 5),
y = rep(seq(-18.01, -18.05, -0.01), each = 5))
template_raster <- Region$new(coordinates = coordinates)$region_raster # full extent
template_raster[][-c(7, 9, 12, 14, 17:19)] <- NA # make U Island
region <- Region$new(template_raster = template_raster)
initial_abundance <- seq(0, 300, 50)
raster::plot(region$raster_from_values(initial_abundance),
main = "Initial abundance", xlab = "Longitude (degrees)",
ylab = "Latitude (degrees)", zlim = c(0, 300), colNA = "blue")
# Set population model
pop_model <- PopulationModel$new(
region = region,
time_steps = 5,
populations = 7,
initial_abundance = initial_abundance,
stage_matrix = matrix(c(0, 2.5, # Leslie/Lefkovitch matrix
0.8, 0.5), nrow = 2, ncol = 2, byrow = TRUE),
carrying_capacity = rep(200, 7),
density_dependence = "logistic",
dispersal = (!diag(nrow = 7, ncol = 7))*0.05,
result_stages = c(1, 2))
# Run single simulation
results <- population_simulator(pop_model)
results # examine
#> $all
#> $all$abundance
#> [1] 1067 1107 1263 1327 1336
#>
#> $all$abundance_stages
#> $all$abundance_stages[[1]]
#> [1] 652 662 754 820 744
#>
#> $all$abundance_stages[[2]]
#> [1] 415 445 509 507 592
#>
#>
#>
#> $abundance
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 52 124 154 177 193
#> [2,] 100 149 195 180 191
#> [3,] 129 153 159 179 173
#> [4,] 152 145 167 168 183
#> [5,] 219 169 202 204 216
#> [6,] 205 172 186 210 180
#> [7,] 210 195 200 209 200
#>
#> $abundance_stages
#> $abundance_stages[[1]]
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 31 73 91 108 103
#> [2,] 67 76 118 110 119
#> [3,] 80 99 103 110 111
#> [4,] 100 74 108 93 99
#> [5,] 138 98 125 118 121
#> [6,] 121 113 106 145 84
#> [7,] 115 129 103 136 107
#>
#> $abundance_stages[[2]]
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 21 51 63 69 90
#> [2,] 33 73 77 70 72
#> [3,] 49 54 56 69 62
#> [4,] 52 71 59 75 84
#> [5,] 81 71 77 86 95
#> [6,] 84 59 80 65 96
#> [7,] 95 66 97 73 93
raster::plot(region$raster_from_values(results$abundance[,5]),
main = "Final abundance", xlab = "Longitude (degrees)",
ylab = "Latitude (degrees)", zlim = c(0, 300), colNA = "blue")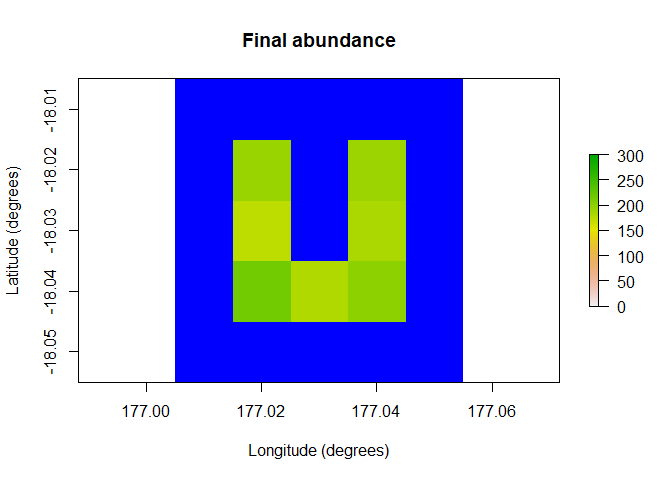
Further examples utilizing the POM workflow and more advanced features of poems can be found in the accompanying vignettes.
Beaumont, M. A., Zhang, W., & Balding, D. J. (2002). ‘Approximate Bayesian computation in population genetics’. Genetics, vol. 162, no. 4, pp, 2025–2035.
Chang, W. (2020). ‘R6: Encapsulated Classes with Reference Semantics’. R package version 2.5.0. Retrieved from https://CRAN.R-project.org/package=R6
Csillery, K., Lemaire L., Francois O., & Blum M. (2015). ‘abc: Tools for Approximate Bayesian Computation (ABC)’. R package version 2.1. Retrieved from https://CRAN.R-project.org/package=abc
Grimm, V., Revilla, E., Berger, U., Jeltsch, F., Mooij, W. M., Railsback, S. F., Thulke, H. H., Weiner, J., Wiegand, T., DeAngelis, D. L., (2005). ‘Pattern-Oriented Modeling of Agent-Based Complex Systems: Lessons from Ecology’. Science vol. 310, no. 5750, pp. 987–991.
Iman R. L., Conover W. J. (1980). ‘Small sample sensitivity analysis techniques for computer models, with an application to risk assessment’. Commun Stat Theor Methods A9, pp. 1749–1842.